Here are the classes, structs, unions and interfaces with brief descriptions:
[detail level 1234]
| ►Neol_bspline | |
| CBSpline | |
| ►NKDTree | |
| ►C_Alloc_base | |
| CNoLeakAlloc | |
| C_Base_iterator | |
| C_Bracket_accessor | |
| C_Iterator | |
| C_Node | |
| C_Node_base | |
| C_Node_compare | |
| C_Region | |
| Calways_true | |
| CKDTree | |
| Csquared_difference | |
| Csquared_difference_counted | |
| ►Nms | |
| ►Nnumpress | |
| ►NOpenMS | Main OpenMS namespace |
| ►NDataArrays | |
| CFloatDataArray | Float data array class |
| CIntegerDataArray | Integer data array class |
| CStringDataArray | String data array class |
| ►NException | Exception namespace |
| CBaseException | Exception base class |
| CBufferOverflow | Buffer overflow exception |
| CConversionError | Invalid conversion exception |
| CDepletedIDPool | Exception used if no more unique document ID's can be drawn from ID pool |
| CDivisionByZero | Division by zero error exception |
| CElementNotFound | Element could not be found exception |
| CFailedAPICall | A call to an external library (other than OpenMS) went wrong |
| CFileEmpty | File is empty |
| CFileNameTooLong | Filename is too long to be writable/readable by the filesystem |
| CFileNotFound | File not found exception |
| CFileNotReadable | File not readable exception |
| CFileNotWritable | File not writable exception |
| CGlobalExceptionHandler | OpenMS global exception handler |
| CIllegalArgument | A method or algorithm argument contains illegal values |
| CIllegalPosition | Invalid 3-dimensional position exception |
| CIllegalSelfOperation | Illegal self operation exception |
| CIllegalTreeOperation | Illegal tree operation exception |
| CIncompatibleIterators | Incompatible iterator exception |
| CIndexOverflow | Int overflow exception |
| CIndexUnderflow | Int underflow exception |
| CInvalidIterator | Invalid iterator exception |
| CInvalidParameter | Exception indicating that an invalid parameter was handed over to an algorithm |
| CInvalidRange | Invalid range exception |
| CInvalidSize | Invalid UInt exception |
| CInvalidValue | Invalid value exception |
| CIOException | General IOException |
| CMissingInformation | Not all required information provided |
| CNotImplemented | Not implemented exception |
| CNullPointer | Null pointer argument is invalid exception |
| COutOfGrid | Out of grid exception |
| COutOfMemory | Out of memory exception |
| COutOfRange | Out of range exception |
| CParseError | Parse Error exception |
| CPostcondition | Postcondition failed exception |
| CPrecondition | Precondition failed exception |
| CRequiredParameterNotGiven | A required parameter was not given |
| CSizeUnderflow | UInt underflow exception |
| CUnableToCalibrate | Exception used if an error occurred while calibrating a dataset |
| CUnableToCreateFile | Unable to create file exception |
| CUnableToFit | Exception used if an error occurred while fitting a model to a given dataset |
| CUnregisteredParameter | An unregistered parameter was accessed |
| CWrongParameterType | A parameter was accessed with the wrong type |
| ►Nims | |
| ►CIMSAlphabet | Holds an indexed list of bio-chemical elements |
| CMassSortingCriteria_ | Private class-functor to sort out elements in mass ascending order |
| CIMSAlphabetParser | An abstract templatized parser to load the data that is used to initialize Alphabet objects |
| CIMSAlphabetTextParser | Implements abstract AlphabetParser to read data from the plain text format |
| CIMSElement | Represents a chemical atom with name and isotope distribution |
| ►CIMSIsotopeDistribution | Represents a distribution of isotopes restricted to the first K elements |
| CPeak | Structure that represents an isotope peak - pair of mass and abundance |
| CIntegerMassDecomposer | Implements MassDecomposer interface using algorithm and data structures described in paper "Efficient Mass Decomposition" S. Böcker, Z. Lipták, ACM SAC-BIO, 2004 doi:10.1145/1066677.1066715 |
| CMassDecomposer | An interface to handle decomposing of integer values/masses over a set of integer weights (alphabet) |
| CRealMassDecomposer | Handles decomposing of non-integer values/masses over a set of non-integer weights with an error allowed |
| CWeights | Represents a set of weights (double values and scaled with a certain precision their integer counterparts) with a quick access |
| ►NInterfaces | |
| CBinaryDataArray | The datastructures used by the OpenSwath interfaces |
| CChromatogram | A single chromatogram |
| CChromatogramMeta | Identifying information for a chromatogram |
| CIChromatogramsReader | The interface of read-access to a list of chromatograms |
| CIChromatogramsWriter | |
| CIMSDataConsumer | The interface of a consumer of spectra and chromatograms |
| CISpectraReader | The interface of read-access to a list of spectra |
| CISpectraWriter | |
| CSpectrum | The structure that captures the generation of a peak list (including the underlying acquisitions) |
| CSpectrumMeta | Identifying information for a spectrum |
| ►NInternal | Namespace used to hide implementation details from users |
| CAcqusHandler | Read-only acqus File handler for XMass Analysis |
| CAreaIterator | Forward iterator for an area of peaks in an experiment |
| CDIntervalBase | A base class for D-dimensional interval |
| CFidHandler | Read-only fid File handler for XMass Analysis |
| CFileMapping | Maps input/output files to filenames for the external program |
| CIntensityIterator | Intensity iterator for a FeatureFinderDefs::IndexSet |
| CIntensityIteratorWrapper | An iterator wrapper to access peak intensities instead of the peak itself |
| CIntensityLess | Comparator that allows to compare the indices of two peaks by their intensity |
| CListEditorDelegate | Internal delegate class |
| CListTable | |
| CMappingParam | Filename mappings for all input/output files |
| CMascotXMLHandler | Handler that is used for parsing MascotXML data |
| CMzDataHandler | |
| CMzDataValidator | Semantically validates MzXML files |
| ►CMzIdentMLDOMHandler | XML DOM handler for MzIdentMLFile |
| CAnalysisSoftware | Struct to hold the used analysis software for that file |
| CDatabaseInput | Struct to hold the information from the DatabaseInput xml tag |
| CDBSequence | Struct to hold the information from the DBSequence xml tag |
| CModificationParam | Struct to hold the information from the ModificationParam xml tag |
| CPeptideEvidence | Struct to hold the PeptideEvidence information |
| CSpectrumIdentification | Struct to hold the information from the SpectrumIdentification xml tag |
| CSpectrumIdentificationProtocol | Struct to hold the information from the SpectrumIdentificationProtocol xml tag |
| CMzIdentMLHandler | XML STREAM handler for MzIdentMLFile |
| CMzIdentMLValidator | Semantically validates MzXML files |
| CMzIterator | M/z iterator for a FeatureFinderDefs::IndexSet |
| ►CMzMLHandler | |
| CChromatogramData | Data necessary to generate a single chromatogram |
| CSpectrumData | Data necessary to generate a single spectrum |
| ►CMzMLHandlerHelper | |
| CBinaryData | Binary data representation |
| CMzMLSqliteHandler | Sqlite handler for storing spectra and chromatograms |
| CMzMLValidator | Semantically validates MzXML files |
| CMzQuantMLHandler | XML handler for MzQuantMLFile |
| CMzQuantMLValidator | Semantically validates MzQuantML files |
| ►CMzXMLHandler | |
| CSpectrumData | Data necessary to generate a single spectrum |
| COpenMSLineEdit | Custom QLineEdit which emits a signal when losing focus (such that we can commit its data) |
| COpenMSOSInfo | |
| CParamEditorDelegate | Internal delegate class for QTreeWidget |
| CParamTree | QTreeWidget that emits a signal whenever a new row is selected |
| CParamXMLHandler | XML Handler for Param files |
| CPTMXMLHandler | Handler that is used for parsing PTMXML data |
| CRtIterator | Retention time iterator for a FeatureFinderDefs::IndexSet |
| ►CSemanticValidator | Semantically validates XML files using CVMappings and a ControlledVocabulary |
| CCVTerm | Representation of a parsed CV term |
| CSpectrum1DPrefDialog | Preferences dialog for Spectrum1DWidget |
| CSpectrum2DPrefDialog | Preferences dialog for Spectrum2DWidget |
| CSpectrum3DPrefDialog | Preferences dialog for Spectrum3DWidget |
| CStringManager | |
| CToolDescription | |
| CToolDescriptionHandler | XML handler for ToolDescriptionFile |
| CToolDescriptionInternal | ToolDescription Class |
| CToolExternalDetails | |
| CTOPPViewPrefDialog | Preferences dialog for TOPPView |
| CTraMLHandler | XML handler for TraMLFile |
| CTraMLValidator | Semantically validates MzXML files |
| CUnimodXMLHandler | Handler that is used for parsing XTandemXML data |
| CXMLFile | Base class for loading/storing XML files that have a handler derived from XMLHandler |
| ►CXMLHandler | Base class for XML handlers |
| CEndParsingSoftly | Exception that is thrown if the parsing is ended by some event (e.g. if only a prefix of the XML file is needed) |
| CXQuestResultXMLHandler | XMLHandler for the result files of XQuest |
| ►NLogger | Log streams |
| CLogStream | Log Stream Class |
| ►CLogStreamBuf | Stream buffer used by LogStream |
| CLogCacheStruct | Holds a counter of occurrences and an index for the occurrence sequence of the corresponding log message |
| CStreamStruct | Holds a stream that is connected to the LogStream. It also includes the minimum and maximum level at which the LogStream redirects messages to this stream |
| CLogStreamNotifier | |
| ►NMath | Math namespace |
| CAsymmetricStatistics | Internal class for asymmetric distributions |
| CAveragePosition | Maintain an average position by summing up positions with weights |
| CBasicStatistics | Calculates some basic statistical parameters of a distribution: sum, mean, variance, and provides the normal approximation |
| CBilinearInterpolation | Provides access to bilinearly interpolated values (and derivatives) from discrete data points. Values beyond the given range of data points are implicitly taken as zero |
| CCumulativeBinomial | Computes the cumulative binomial function |
| ►CGammaDistributionFitter | Implements a fitter for the Gamma distribution |
| CGammaDistributionFitResult | Struct to represent the parameters of a gamma distribution |
| ►CGaussFitter | Implements a fitter for Gaussian functions |
| CGaussFitResult | Struct of parameters of a Gaussian distribution |
| ►CGumbelDistributionFitter | Implements a fitter for the Gumbel distribution |
| CGumbelDistributionFitResult | Struct to represent the parameters of a gumbel distribution |
| CHistogram | Representation of a histogram |
| CLinearInterpolation | Provides access to linearly interpolated values (and derivatives) from discrete data points. Values beyond the given range of data points are implicitly taken as zero |
| CLinearRegression | This class offers functions to perform least-squares fits to a straight line model, 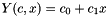 |
| CPosteriorErrorProbabilityModel | Implements a mixture model of the inverse gumbel and the gauss distribution or a gaussian mixture |
| CQuadraticRegression | |
| CRANSAC | This class provides a generic implementation of the RANSAC outlier detection algorithm. Is implemented and tested after the SciPy reference: http://wiki.scipy.org/Cookbook/RANSAC |
| CRansacModel | Generic plug-in template base class using 'Curiously recurring template pattern' (CRTP) to allow for arbitrary RANSAC models (e.g. linear or quadratic fits) |
| CRansacModelLinear | Implementation of a linear RANSAC model fit |
| CRansacModelQuadratic | Implementation of a quadratic RANSAC model fit |
| CRANSACParam | A simple struct to carry all the parameters required for a RANSAC run |
| ►CROCCurve | ROCCurves show the trade-off in sensitivity and specificity for binary classifiers using different cutoff values |
| Csimsortdec | Predicate for sort() |
| CSummaryStatistics | Helper class to gather (and dump) some statistics from a e.g. vector<double> |
| ►NOptimizationFunctions | |
| CPenaltyFactors | Class for the penalty factors used during the optimization |
| CPenaltyFactorsIntensity | Class for the penalty factors used during the optimization |
| ►NSimTypes | |
| CSimProtein | Plain data object holding sequence and abundance information on a single protein |
| CSimRandomNumberGenerator | Wrapper class for random number generators used by the simulation classes |
| ►NTargetedExperimentHelper | This class stores helper structures that are used in multiple classes of the TargetedExperiment (e.g. ReactionMonitoringTransition and IncludeExcludeTarget) |
| CCompound | |
| CConfiguration | |
| CContact | |
| CCV | |
| CInstrument | |
| CInterpretation | |
| ►CPeptide | |
| CModification | |
| CPrediction | |
| CProtein | |
| CPublication | |
| CRetentionTime | |
| CTraMLProduct | |
| CAAIndex | Representation of selected AAIndex properties |
| ►CAASequence | Representation of a peptide/protein sequence |
| CConstIterator | ConstIterator for AASequence |
| CIterator | Iterator class for AASequence |
| ►CAccurateMassSearchEngine | An algorithm to search for exact mass matches from a spectrum against a database (e.g. HMDB) |
| CCompareEntryAndMass_ | |
| CMappingEntry_ | |
| CAccurateMassSearchResult | |
| CAcquisition | Information about one raw data spectrum that was combined with several other raw data spectra |
| CAcquisitionInfo | Description of the combination of raw data to a single spectrum |
| CAcquisitionInfoVisualizer | Class that displays all meta information for AcquisitionInfo objects |
| CAcquisitionVisualizer | Class that displays all meta information for Acquisition objects |
| CAdduct | |
| CAdductInfo | |
| CAnnotation1DCaret | An annotation item which paints a set of carets on the canvas |
| CAnnotation1DDistanceItem | An annotation item which represents a measured distance between two peaks |
| CAnnotation1DItem | An abstract class acting as an interface for the different 1D annotation items |
| CAnnotation1DPeakItem | A peak annotation item |
| CAnnotation1DTextItem | An annotation item which represents an arbitrary text on the canvas |
| CAnnotations1DContainer | Container for annotations to content of Spectrum1DCanvas |
| CAnnotationStatistics | |
| CAScore | Implementation of the Ascore For a given peptide sequence and its MS/MS spectrum it identifies the most probable phosphorylation-site(s). For each phosphorylation site a probability score is calculated. The algorithm is implemented according to Beausoleil et al |
| CAverageLinkage | AverageLinkage ClusterMethod |
| CAxisPainter | Draws a coordinate axis. It has only static methods, that's why the constructor is private |
| CAxisTickCalculator | Calculates ticks for a given value range |
| CAxisWidget | Widget that represents an axis of a graph |
| CBackgroundControl | |
| CBackgroundIntensityBin | |
| ►CBase64 | Class to encode and decode Base64 |
| CReinterpreter32_ | Internal class needed for type-punning |
| CReinterpreter64_ | Internal class needed for type-punning |
| ►CBaseFeature | A basic LC-MS feature |
| CQualityLess | Compare by quality |
| CBaseGroupFinder | The base class of all element group finding algorithms |
| CBaseLabeler | Abstract base class for all kinds of labeling techniques |
| CBaseModel | Abstract base class for all D-dimensional models |
| CBaseSuperimposer | The base class of all superimposer algorithms |
| CBaseVisualizer | A base class for all visualizer classes |
| CBaseVisualizerGUI | A base class for all visualizer classes |
| CBernNorm | BernNorm scales the peaks by ranking them and then scaling them according to rank |
| CBiGaussFitter1D | BiGaussian distribution fitter (1-dim.) approximated using linear interpolation |
| CBiGaussModel | BiGaussian distribution approximated using linear interpolation |
| CBigString | Concatenates Proteins given as FASTAEntry to one big string separated by a unique character (by default $) |
| CBinaryComposeFunctionAdapter | Represents the binary compose function object adapter |
| CBinaryTreeNode | Elements of a binary tree used to represent a hierarchical clustering process |
| CBinnedSharedPeakCount | Compare functor scoring the shared peaks for similarity measurement |
| CBinnedSpectralContrastAngle | Compare functor scoring the spectral contrast angle for similarity measurement |
| ►CBinnedSpectrum | This is a binned representation of a PeakSpectrum |
| CNoSpectrumIntegrated | Exception which is thrown if BinnedSpectrum bins are accessed and no PeakSpektrum has been integrated yet i.e. bins_ is empty |
| ►CBinnedSpectrumCompareFunctor | Base class for compare functors of BinnedSpectra |
| CIncompatibleBinning | Exception thrown if compared spectra are incompatible |
| CBinnedSumAgreeingIntensities | Compare functor scoring the sum of agreeing intensities for similarity measurement |
| CBSpline2d | B spline interpolation |
| CBzip2Ifstream | Decompresses files which are compressed in the bzip2 format (*.bz2) |
| CBzip2InputStream | Implements the BinInputStream class of the xerces-c library in order to read bzip2 compressed XML files |
| CCachedmzML | An class that uses on-disk caching to read and write spectra and chromatograms |
| CCachedSwathFileConsumer | On-disk cached implementation of FullSwathFileConsumer |
| CCalibrationData | A helper class, holding all calibration points |
| CCentroidData | |
| CCentroidPeak | |
| CChargePair | Representation of a (putative) link between two Features, which stem from the same compound but have different charge (including different adduct ions (H+, Na+, ..) |
| CChromatogramExtractor | The ChromatogramExtractor extracts chromatograms from a spectra file |
| ►CChromatogramExtractorAlgorithm | The ChromatogramExtractorAlgorithm extracts chromatograms from a MS data |
| CExtractionCoordinates | |
| ►CChromatogramPeak | A 1-dimensional raw data point or peak for chromatograms |
| CIntensityLess | Comparator by intensity |
| CPositionLess | Comparator by position. As this class has dimension 1, this is basically an alias for RTLess |
| CRTLess | Comparator by RT position |
| CChromatogramSettings | Representation of chromatogram settings, e.g. SRM/MRM chromatograms |
| CChromatogramTools | Conversion class to convert chromatograms |
| CChromExtractParams | ChromatogramExtractor parameters |
| CClusterAnalyzer | Bundles analyzing tools for a clustering (given as sequence of BinaryTreeNode's) |
| CClusteredMS2ConsensusSpectrum | |
| ►CClusterFunctor | Base class for cluster functors |
| CInsufficientInput | Exception thrown if not enough data (<2) is used |
| CClusterHierarchical | Hierarchical clustering with generic clustering functions |
| CClusteringGrid | Data structure to store 2D data to be clustered e.g. (m/z, retention time) coordinates from multiplex filtering |
| CClusterProxyKD | Proxy for a (potential) cluster |
| CCmpHypothesesByScore | |
| CCmpMassTraceByMZ | |
| CColorSelector | A widget for selecting a color |
| CComplementFilter | Total intensity of peak pairs that could result from complementing fragments of charge state 1 |
| CComplementMarker | ComplementMarker marks peak pairs which could represent y - b ion pairs |
| CCompleteLinkage | CompleteLinkage ClusterMethod |
| CCompNovoIdentification | Run with CompNovoIdentification |
| ►CCompNovoIdentificationBase | Run with CompNovoIdentificationBase |
| CPermut | Simple class to store permutations and a score |
| CCompNovoIdentificationCID | Run with CompNovoIdentificationCID |
| CCompNovoIonScoring | Run with CompNovoIonScoring |
| ►CCompNovoIonScoringBase | Run with CompNovoIonScoringBase |
| CIonScore | |
| CCompNovoIonScoringCID | Run with CompNovoIonScoringCID |
| CCompomer | Holds information on an edge connecting two features from a (putative) charge ladder |
| CCompressedInputSource | This class is based on xercesc::LocalFileInputSource |
| CConfidenceScoring | |
| CConnectedComponent | |
| ►CConsensusFeature | A 2-dimensional consensus feature |
| CMapsLess | Compare by the sets of consensus elements (lexicographically) |
| CRatio | Slim struct to feed the need for systematically storing of ratios ( |
| CSizeLess | Compare by size(), the number of consensus elements |
| CConsensusIDAlgorithm | Abstract base class for all ConsensusID algorithms (that calculate a consensus from multiple ID runs) |
| CConsensusIDAlgorithmAverage | Calculates a consensus from multiple ID runs by averaging the search scores |
| CConsensusIDAlgorithmBest | Calculates a consensus from multiple ID runs by taking the best search score |
| CConsensusIDAlgorithmIdentity | Abstract base class for ConsensusID algorithms that compare only identical sequences |
| CConsensusIDAlgorithmPEPIons | Calculates a consensus from multiple ID runs based on PEPs and shared ions |
| CConsensusIDAlgorithmPEPMatrix | Calculates a consensus from multiple ID runs based on PEPs and sequence similarities |
| CConsensusIDAlgorithmRanks | Calculates a consensus from multiple ID runs based on the ranks of the search hits |
| CConsensusIDAlgorithmSimilarity | Abstract base class for ConsensusID algorithms that take peptide similarity into account |
| CConsensusIDAlgorithmWorst | Calculates a consensus from multiple ID runs by taking the worst search score (conservative approach) |
| CConsensusIsotopePattern | |
| ►CConsensusMap | A container for consensus elements |
| CFileDescription | Source file description for input files |
| CConsensusMapNormalizerAlgorithmMedian | Algorithms of ConsensusMapNormalizer |
| CConsensusMapNormalizerAlgorithmQuantile | Algorithms of ConsensusMapNormalizer |
| CConsensusMapNormalizerAlgorithmThreshold | Algorithms of ConsensusMapNormalizer |
| CConsensusXMLFile | This class provides Input functionality for ConsensusMaps and Output functionality for alignments and quantitation |
| CConsoleUtils | |
| ►CConstRefVector | This vector holds pointer to the elements of another container |
| CConstRefVectorConstIterator | ConstIterator for the ConstRefVector |
| CConstRefVectorIterator | Mutable iterator for the ConstRefVector |
| CContactPerson | Contact person information |
| CContactPersonVisualizer | Class that displays all meta information for ContactPerson objects |
| CContinuousWaveletTransform | This class is the base class of the continuous wavelet transformation |
| CContinuousWaveletTransformNumIntegration | This class computes the continuous wavelet transformation using a marr wavelet |
| ►CControlledVocabulary | Representation of a controlled vocabulary |
| CCVTerm | Representation of a CV term |
| CConvexHull2D | A 2-dimensional hull representation in [counter]clockwise direction - depending on axis labelling |
| CCrossLinksDB | |
| ►CCsiFingerIdMzTabWriter | |
| CCsiAdapterHit | Internal structure used in SiriusAdapter that is used for the conversion of the Csi:FingerID output to an mzTab |
| CCsiAdapterIdentification | |
| CCsiAdapterRun | |
| CCsvFile | This class handles csv files. Currently only loading is implemented |
| CCubicSpline2d | Cubic spline interpolation as described in R.L. Burden, J.D. Faires, Numerical Analysis, 4th ed. PWS-Kent, 1989, ISBN 0-53491-585-X, pp. 126-131 |
| CCVMappingFile | Used to load CvMapping files |
| CCVMappingRule | Representation of a CV Mapping rule used by CVMappings |
| CCVMappings | Representation of controlled vocabulary mapping rules (for PSI formats) |
| CCVMappingTerm | Representation of controlled vocabulary term |
| CCVReference | Controlled Vocabulary Reference |
| ►CCVTerm | Representation of controlled vocabulary term |
| CUnit | |
| CCVTermList | Representation of controlled vocabulary term list |
| CCVTermListInterface | Interface to the controlled vocabulary term list |
| CDataFilterDialog | Dialog for creating and changing a DataFilter |
| ►CDataFilters | DataFilter array providing some convenience functions |
| CDataFilter | Representation of a peak/feature filter combining FilterType, FilterOperation and a value |
| CDataProcessing | Description of the applied preprocessing steps |
| CDataProcessingVisualizer | Class that displays all meta information for DataProcessing objects |
| CDataValue | Class to hold strings, numeric values, lists of strings and lists of numeric values |
| CDate | Date Class |
| CDateTime | DateTime Class |
| CDBoundingBox | A D-dimensional bounding box |
| CDeconvPeak | |
| CDefaultParamHandler | A base class for all classes handling default parameters |
| CDeisotoper | |
| CDeNovoAlgorithm | Base class for ion scoring implementation for de novo algorithms |
| CDeNovoIdentification | Base class for de novo identification |
| ►CDeNovoIonScoring | Base class for ion scoring implementation for de novo algorithms |
| CIonScore | IonScore |
| CDeNovoPostScoring | Base class for ion scoring implementation for de novo algorithms |
| CDetectabilitySimulation | Simulates peptide detectability |
| CDiaPrescore | Scoring of an spectrum given library intensities of a transition group |
| CDIAScoring | Scoring of an spectrum at the peak apex of an chromatographic elution peak |
| CDigestion | Meta information about digestion of a sample |
| CDigestionVisualizer | Class that displays all meta information of digestion objects |
| CDigestSimulation | Simulates protein digestion |
| CDistanceMatrix | A two-dimensional distance matrix, similar to OpenMS::Matrix |
| CDocumentIdentifier | Manage source document information |
| CDocumentIdentifierVisualizer | Class that displays all meta information for DocumentIdentifier objects |
| CDocumentIDTagger | Tags OpenMS file containers with a DocumentID |
| CDPeak | Metafunction to choose among Peak1D respectively Peak2D through a template argument |
| CDPosition | Representation of a coordinate in D-dimensional space |
| CDRange | A D-dimensional half-open interval |
| CDTA2DFile | DTA2D File adapter |
| CDTAFile | File adapter for DTA files |
| CEDTAFile | File adapter for Enhanced DTA files |
| ►CEGHFitter1D | Exponential-Gaussian hybrid distribution fitter (1-dim.) using Levenberg-Marquardt algorithm (Eigen implementation) for parameter optimization |
| CData | Helper struct (contains the size of an area and a raw data container) |
| CEGHFitterFunctor | |
| CEGHModel | Exponential-Gaussian hybrid distribution model for elution profiles |
| ►CEGHTraceFitter | A RT Profile fitter using an Exponential Gaussian Hybrid background model |
| CEGHTraceFunctor | |
| CElement | Representation of an element |
| CElementDB | Stores elements |
| CElutionModelFitter | Helper class for fitting elution models to features |
| CElutionPeakDetection | Extracts chromatographic peaks from a mass trace |
| ►CEmgFitter1D | Exponentially modified gaussian distribution fitter (1-dim.) using Levenberg-Marquardt algorithm (Eigen implementation) for parameter optimization |
| CData | Helper struct (contains the size of an area and a raw data container) |
| CEgmFitterFunctor | |
| CEmgModel | Exponentially modified gaussian distribution model for elution profiles |
| CEmgScoring | Scoring of an elution peak using an exponentially modified gaussian distribution model |
| CEmpiricalFormula | Representation of an empirical formula |
| CEnhancedTabBar | Convenience tab bar implementation |
| CEnhancedTabBarWidgetInterface | Widgets that are placed into an EnhancedTabBar must implement this interface |
| CEnhancedWorkspace | |
| CEnzymaticDigestion | Class for the enzymatic digestion of proteins |
| ►CEnzymaticDigestionLogModel | Class for the Log L model of enzymatic digestion of proteins |
| CBindingSite_ | |
| CCleavageModel_ | |
| CEnzyme | Representation of an enzyme |
| CEnzymesDB | Enzyme database which holds enzymes |
| CEqualInTolerance | Struct for binary predicate to consider equality with a certain tolerance |
| CEuclideanSimilarity | CompareFunctor for 2Dpoints |
| ►CExperimentalDesign | A TSV and user friendly representation of the experimental design. Used for loading and storing the experimental design in OpenMS |
| CMSRun | |
| CExperimentalSettings | Description of the experimental settings |
| CExperimentalSettingsVisualizer | Class that displays all meta information for ExperimentalSettings objects |
| CExtendedIsotopeFitter1D | Extended isotope distribution fitter (1-dim.) approximated using linear interpolation |
| CExtendedIsotopeModel | Extended isotope distribution approximated using linear interpolation |
| CFactory | Returns FactoryProduct* based on the name of the desired concrete FactoryProduct |
| CFactoryBase | Base class for Factory<T> |
| CFakeProcess | A FakeProcess class |
| CFalseDiscoveryRate | Calculates an FDR from identifications |
| ►CFASTAFile | This class serves for reading in and writing FASTA files |
| CFASTAEntry | FASTA entry type (identifier, description and sequence) |
| CFastaIterator | Iterator over FASTA file |
| CFastaIteratorIntern | Iterator for a FASTA file |
| CFeaFiModule | Implements a module of the FeatureFinder algorithm |
| CFeature | An LC-MS feature |
| CFeatureDeconvolution | An algorithm to decharge features (i.e. as found by FeatureFinder) |
| ►CFeatureDistance | A functor class for the calculation of distances between features or consensus features |
| CDistanceParams_ | Structure for storing distance parameters |
| CFeatureEditDialog | Dialog for editing a feature |
| CFeatureFileOptions | Options for loading files containing features |
| CFeatureFinder | The main feature finder class |
| CFeatureFinderAlgorithm | Abstract base class for FeatureFinder algorithms |
| ►CFeatureFinderAlgorithmIsotopeWavelet | Implements the isotope wavelet feature finder |
| CBoxElement | Internally used data structure for the sweep line algorithm |
| CFeatureFinderAlgorithmMRM | FeatureFinderAlgorithm for MRM experiments |
| CFeatureFinderAlgorithmPicked | FeatureFinderAlgorithm for picked peaks |
| ►CFeatureFinderAlgorithmPickedHelperStructs | Wrapper struct for all the classes needed by the FeatureFinderAlgorithmPicked and the associated classes |
| CIsotopePattern | Helper structure for a found isotope pattern used in FeatureFinderAlgorithmPicked |
| CMassTrace | Helper struct for mass traces used in FeatureFinderAlgorithmPicked |
| CMassTraces | Helper struct for a collection of mass traces used in FeatureFinderAlgorithmPicked |
| CSeed | Helper structure for seeds used in FeatureFinderAlgorithmPicked |
| CTheoreticalIsotopePattern | Helper structure for a theoretical isotope pattern used in FeatureFinderAlgorithmPicked |
| CFeatureFinderAlgorithmSH | The Superhirn FeatureFinderAlgorithm |
| CFeatureFinderAlgorithmSHCtrl | A facade for various Superhirn FeatureFinder classes. Use FeatureFinderAlgorithmSH instead |
| ►CFeatureFinderDefs | The purpose of this struct is to provide definitions of classes and typedefs which are used throughout all FeatureFinder classes |
| CNoSuccessor | Exception that is thrown if a method an invalid IndexPair is given |
| CFeatureFindingMetabo | Method for the assembly of mass traces belonging to the same isotope pattern, i.e., that are compatible in retention times, mass-to-charge ratios, and isotope abundances |
| CFeatureGroupingAlgorithm | Base class for all feature grouping algorithms |
| CFeatureGroupingAlgorithmKD | A feature grouping algorithm for unlabeled data |
| CFeatureGroupingAlgorithmLabeled | A map feature grouping algorithm for labeling techniques with two labels |
| CFeatureGroupingAlgorithmQT | A feature grouping algorithm for unlabeled data |
| CFeatureGroupingAlgorithmUnlabeled | A map feature grouping algorithm for unlabeled data |
| ►CFeatureHandle | Representation of a Peak2D, RichPeak2D or Feature |
| CFeatureHandleMutable_ | Helper class returned by FeatureHandle::asMutable(), which see |
| CIndexLess | Comparator by map and unique id |
| CFeatureHypothesis | Internal structure used in FeatureFindingMetabo that keeps track of a feature hypothesis (isotope group hypothesis) |
| CFeatureLCProfile | |
| CFeatureMap | A container for features |
| CFeatureOpenMS | An implementation of the OpenSWATH Feature Access interface using OpenMS |
| CFeatureXMLFile | This class provides Input/Output functionality for feature maps |
| ►CFile | Basic file handling operations |
| CTemporaryFiles_ | Internal helper class, which holds temporary filenames and deletes these file at program exit |
| CFileHandler | Facilitates file handling by file type recognition |
| CFileTypes | Centralizes the file types recognized by FileHandler |
| CFileWatcher | Watcher that monitors file changes |
| CFilterFunctor | A FilterFunctor extracts some spectrum characteristics for quality assessment |
| CFitter1D | Abstract base class for all 1D-dimensional model fitter |
| CFTPeakDetectController | |
| CFullSwathFileConsumer | Abstract base class which can consume spectra coming from SWATH experiment stored in a single file |
| ►CFuzzyStringComparator | Fuzzy comparison of strings, tolerates numeric differences |
| CAbortComparison | Internal exception class |
| CInputLine | Stores information about the current input line (i.e., stream for the line and the current position in the stream) |
| CPrefixInfo_ | Wrapper for the prefix information computed for the failure report |
| CStreamElement_ | Stores information about characters, numbers, and white spaces loaded from the InputStream |
| CGaussFilter | This class represents a Gaussian lowpass-filter which works on uniform as well as on non-uniform profile data |
| CGaussFilterAlgorithm | This class represents a Gaussian lowpass-filter which works on uniform as well as on non-uniform profile data |
| CGaussFitter1D | Gaussian distribution fitter (1-dim.) approximated using linear interpolation |
| CGaussModel | Normal distribution approximated using linear interpolation |
| ►CGaussTraceFitter | Fitter for RT profiles using a Gaussian background model |
| CGaussTraceFunctor | |
| CGoodDiffFilter | GoodDiffFilter counts the number ob peak pairs whose m/z difference can be explained by a amino acid loss |
| CGradient | Representation of a HPLC gradient |
| CGradientVisualizer | GradientVisualizer is a visualizer class for objects of type gradient |
| CGridBasedCluster | Basic data structure for clustering |
| CGridBasedClustering | 2D hierarchical clustering implementation optimized for large data sets containing many small clusters i.e. dimensions of clusters << dimension of entire dataset |
| CGridFeature | Representation of a feature in a hash grid |
| CGUIHelpers | Class which holds static GUI-related helper functions |
| CGUIProgressLoggerImpl | Implements a GUI version of the ProgressLoggerImpl |
| CGzipIfstream | Decompresses files which are compressed in the gzip format (*.gzip) |
| CGzipInputStream | Implements the BinInputStream class of the xerces-c library in order to read gzip compressed XML files |
| CHasActivationMethod | Predicate that determines if a spectrum was generated using any activation method given in the constructor list |
| ►CHashGrid | Container for (2-dimensional coordinate, value) pairs |
| CConstIterator | Constant element iterator for the hash grid |
| CIterator | Element iterator for the hash grid |
| CHasMetaValue | Predicate that determines if a class has a certain metavalue |
| CHasPrecursorCharge | Predicate that determines if a spectrum has a certain precursor charge as given in the constructor list |
| CHasScanMode | Predicate that determines if a spectrum has a certain scan mode |
| CHasScanPolarity | Predicate that determines if a spectrum has a certain scan polarity |
| CHiddenMarkovModel | Hidden Markov Model implementation of PILIS |
| CHistogramDialog | Dialog that show a HistogramWidget |
| CHistogramWidget | Widget which can visualize a histogram |
| CHMMState | Hidden Markov Model State class for the Hidden Markov Model |
| CHPLC | Representation of a HPLC experiment |
| CHPLCVisualizer | Class that displays all meta information for HPLC objects |
| CHyperScore | An implementation of the X!Tandem HyperScore PSM scoring function |
| CIBSpectraFile | Implements the export of consensusmaps into the IBSpectra format used by isobar to load quantification results |
| CICPLLabeler | Simulate ICPL experiments |
| ►CIDDecoyProbability | IDDecoyProbability calculates probabilities using decoy approach |
| CTransformation_ | Struct to be used to store a transformation (used for fitting) |
| CIdentification | Represents a object which can store the information of an analysisXML instance |
| CIdentificationHit | Represents a object which can store the information of an analysisXML instance |
| CIDEvaluationBase | Main window of the IDEvaluation tool |
| ►CIDFilter | Collection of functions for filtering peptide and protein identifications |
| CDigestionFilter | Is peptide evidence digestion product of some protein |
| CGetMatchingItems | Builds a map index of data that have a String index to find matches and return the objects |
| CHasDecoyAnnotation | Is this a decoy hit? |
| CHasGoodScore | Is the score of this hit at least as good as the given value? |
| CHasMatchingAccession | Given a list of protein accessions, do any occur in the annotation(s) of this hit? |
| CHasMaxMetaValue | Does a meta value of this hit have at most the given value? |
| CHasMaxRank | Is the rank of this hit below or at the given cut-off? |
| CHasMetaValue | Is a meta value with given key and value set on this hit? |
| CHasNoHits | Is the list of hits of this peptide/protein ID empty? |
| ►CIDMapper | Annotates an MSExperiment, FeatureMap or ConsensusMap with peptide identifications |
| CSpectraIdentificationState | Result of a partitioning by identification state with mapPrecursorsToIdentifications() |
| CIDRipper | Ripping protein/peptide identification according their file origin |
| CIdXMLFile | Used to load and store idXML files |
| CILPDCWrapper | |
| CIncludeExcludeTarget | This class stores a SRM/MRM transition |
| ►CInclusionExclusionList | Provides functionality for writing inclusion or exclusion lists |
| CIEWindow | |
| CWindowDistance_ | Determine distance between two spectra |
| CIndexedMzMLDecoder | A class to analyze indexedmzML files and extract the offsets of individual tags |
| CIndexedMzMLFile | A low-level class to read an indexedmzML file |
| CIndexedMzMLFileLoader | A class to load an indexedmzML file |
| CINIFileEditorWindow | Shows the ParamEditor widget in a QMainWindow with a toolbar |
| CInIntensityRange | Predicate that determines if a peak lies inside/outside a specific intensity range |
| CINIUpdater | |
| CInMSLevelRange | Predicate that determines if a spectrum lies inside/outside a specific MS level set |
| CInMzRange | Predicate that determines if a peak lies inside/outside a specific m/z range |
| CInPrecursorMZRange | Predicate that determines if a spectrum's precursor is within a certain m/z range |
| CInRTRange | Predicate that determines if a spectrum lies inside/outside a specific retention time range |
| CInspectInfile | Inspect input file adapter |
| CInspectOutfile | Representation of an Inspect outfile |
| CInstrument | Description of a MS instrument |
| CInstrumentSettings | Description of the settings a MS Instrument was run with |
| CInstrumentSettingsVisualizer | Class that displays all meta information for InstrumentSettings objects |
| CInstrumentVisualizer | Class that displays all meta information for an MS instrument |
| CIntensityBalanceFilter | IntensityBalanceFilter divides the m/z-range into ten regions and sums the intensity in these regions |
| ►CInternalCalibration | A mass recalibration method using linear/quadratic interpolation (robust/weighted) of given reference masses |
| CLockMass | Helper class, describing a lock mass |
| CInterpolationModel | Abstract class for 1D-models that are approximated using linear interpolation |
| CIonDetector | Description of a ion detector (part of a MS Instrument) |
| CIonDetectorVisualizer | Class that displays all meta information for IonDetector objects |
| CIonizationSimulation | Simulates Protein ionization |
| CIonSource | Description of an ion source (part of a MS Instrument) |
| CIonSourceVisualizer | Class that displays all meta information for IonSource objects |
| CIsEmptySpectrum | Predicate that determines if a spectrum is empty |
| CIsInCollisionEnergyRange | Predicate that determines if an MSn spectrum was generated with a collision energy in the given range |
| CIsInIsolationWindow | Predicate that determines if the isolation window covers ANY of the given m/z values |
| CIsInIsolationWindowSizeRange | Predicate that determines if the width of the isolation window of an MSn spectrum is in the given range |
| ►CIsobaricChannelExtractor | Extracts individual channels from MS/MS spectra for isobaric labeling experiments |
| CPuritySate_ | Small struct to capture the current state of the purity computation |
| CIsobaricIsotopeCorrector | Performs isotope impurity correction on the intensities extracted from an isobaric labeling experiment |
| CIsobaricNormalizer | Performs median normalization on the extracted ratios of isobaric labeling experiment |
| CIsobaricQuantifier | Given the extracted channel intensities the IsobaricQuantifier corrects and normalizes the intensities for further processing |
| CIsobaricQuantifierStatistics | Statistics for quantitation performance and comparison of NNLS vs. naive method (aka matrix inversion) |
| ►CIsobaricQuantitationMethod | Abstract base class describing an isobaric quantitation method in terms of the used channels and an isotope correction matrix |
| CIsobaricChannelInformation | Summary of an isobaric quantitation channel |
| ►CIsotopeCluster | Stores information about an isotopic cluster (i.e. potential peptide charge variants) |
| CChargedIndexSet | Index set with associated charge estimate |
| CIsotopeDiffFilter | IsotopeDiffFilter returns total intensity of peak pairs that could result from isotope peaks |
| CIsotopeDistribution | Isotope distribution class |
| CIsotopeDistributionCache | Pre-calculate isotope distributions for interesting mass ranges |
| CIsotopeFitter1D | Isotope distribution fitter (1-dim.) approximated using linear interpolation |
| CIsotopeMarker | IsotopeMarker marks peak pairs which could represent an ion and its isotope |
| CIsotopeModel | Isotope distribution approximated using linear interpolation |
| ►CIsotopeWavelet | Implements the isotope wavelet function |
| Cfi_ | Internal union for fast computation of the power function |
| ►CIsotopeWaveletTransform | A class implementing the isotope wavelet transform. If you just want to find features using the isotope wavelet, take a look at the FeatureFinderAlgorithmIsotopeWavelet class. Usually, you only have to consider the class at hand if you plan to change the basic implementation of the transform |
| CBoxElement | Internally used data structure |
| CTransSpectrum | Internally (only by GPUs) used data structure . It allows efficient data exchange between CPU and GPU and avoids unnecessary memory moves. The class is tailored on the isotope wavelet transform and is in general not applicable on similar - but different - situations |
| CIsotopicDist | |
| CIsZoomSpectrum | Predicate that determines if a spectrum is a zoom (enhanced resolution) spectrum |
| ►CItraqConstants | Some constants used throughout iTRAQ classes |
| CChannelInfo | Stores information on an iTRAQ channel |
| CItraqEightPlexQuantitationMethod | ITRAQ 8 plex quantitation to be used with the IsobaricQuantitation |
| CItraqFourPlexQuantitationMethod | ITRAQ 4 plex quantitation to be used with the IsobaricQuantitation |
| CITRAQLabeler | Simulate iTRAQ experiments |
| CJavaInfo | Detect Java and retrieve information |
| CKDTreeFeatureMaps | Stores a set of features, together with a 2D tree for fast search |
| CKDTreeFeatureNode | A node of the kD-tree with pointer to corresponding data and index |
| CKroenikFile | File adapter for Kroenik (HardKloer sibling) files |
| CLabeledPairFinder | The LabeledPairFinder allows the matching of labeled features (features with a fixed distance) |
| CLabelFreeLabeler | Abstract base class for all kinds of labeling techniques |
| CLayerData | Class that stores the data for one layer |
| ►CLayerStatisticsDialog | Dialog showing statistics about the data of the current layer |
| CMetaStatsValue_ | Struct representing the statistics about one meta information |
| CLCElutionPeak | |
| ►CLCMS | |
| COPERATOR_FeatureCompare | |
| COPERATOR_MZ | |
| CLCMSCData | |
| ►CLevMarqFitter1D | Abstract class for 1D-model fitter using Levenberg-Marquardt algorithm for parameter optimization |
| CGenericFunctor | |
| CLexicographicComparator | A wrapper class that combines two comparators lexicographically. Normally you should use the make-function lexicographicComparator() because then you do not need to specify the template arguments |
| CLibSVMEncoder | Serves for encoding sequences into feature vectors |
| CLinearResampler | Linear Resampling of raw data |
| CLinearResamplerAlign | Linear Resampling of raw data with alignment |
| CListEditor | Editor for editing int, double and string lists (including output and input file lists) |
| ►CListUtils | Collection of utility functions for management of vectors |
| CDoubleTolerancePredicate_ | Predicate to check double equality with a given tolerance |
| ►CLocalLinearMap | Trained Local Linear Map (LLM) model for peak intensity prediction |
| CLLMParam | Define parameters needed by the Local Linear Map (LLM) model |
| CLogConfigHandler | The LogConfigHandler provides the functionality to configure the internal logging of OpenMS algorithms that use the global instances of LogStream |
| CLowessSmoothing | LOWESS (locally weighted scatterplot smoothing) |
| ►CLPWrapper | |
| CSolverParam | Struct that holds the parameters of the LP solver |
| ►CMap | Map class based on the STL map (containing several convenience functions) |
| CIllegalKey | Map illegal key exception |
| CMapAlignmentAlgorithmIdentification | A map alignment algorithm based on peptide identifications from MS2 spectra |
| CMapAlignmentAlgorithmKD | An efficient reference-free feature map alignment algorithm for unlabeled data |
| CMapAlignmentAlgorithmPoseClustering | A map alignment algorithm based on pose clustering |
| ►CMapAlignmentAlgorithmSpectrumAlignment | A map alignment algorithm based on spectrum similarity (dynamic programming) |
| CCompare | Inner class necessary for using the sort algorithm |
| CMapAlignmentEvaluationAlgorithm | Base class for all Caap evaluation algorithms |
| CMapAlignmentEvaluationAlgorithmPrecision | Caap evaluation algorithm to obtain a precision value |
| CMapAlignmentEvaluationAlgorithmRecall | Caap evaluation algorithm to obtain a recall value |
| CMapAlignmentTransformer | This class collects functions for applying retention time transformations to data structures |
| CMapConversion | |
| CMarkerMower | MarkerMower uses PeakMarker to find peaks, those that are not marked get removed |
| CMascotGenericFile | Read/write Mascot generic files (MGF) |
| CMascotInfile | Mascot input file adapter |
| CMascotRemoteQuery | Class which handles the communication between OpenMS and the Mascot server |
| CMascotXMLFile | Used to load Mascot XML files |
| CMassAnalyzer | Description of a mass analyzer (part of a MS Instrument) |
| CMassAnalyzerVisualizer | Class that displays all meta information for MassAnalyzer objects |
| CMassDecomposition | Class represents a decomposition of a mass into amino acids |
| CMassDecompositionAlgorithm | Mass decomposition algorithm, given a mass it suggests possible compositions |
| CMassExplainer | Computes empirical formulas for given mass differences using a set of allowed elements |
| CMassTrace | A container type that gathers peaks similar in m/z and moving along retention time |
| CMassTraceDetection | A mass trace extraction method that gathers peaks similar in m/z and moving along retention time |
| CMatrix | A two-dimensional matrix. Similar to std::vector, but uses a binary operator(,) for element access |
| CMaxLikeliFitter1D | Abstract base class for all 1D-model fitters using maximum likelihood optimization |
| CMetaboliteFeatureDeconvolution | An algorithm to decharge small molecule features (i.e. as found by FeatureFinder) |
| CMetaboliteSpectralMatching | |
| CMetaDataBrowser | A meta data visualization widget |
| CMetaInfo | A Type-Name-Value tuple class |
| CMetaInfoDescription | Description of the meta data arrays of MSSpectrum |
| CMetaInfoDescriptionVisualizer | Class that displays all meta information for MetaInfoDescription objects |
| CMetaInfoInterface | Interface for classes that can store arbitrary meta information (Type-Name-Value tuples) |
| CMetaInfoInterfaceUtils | Utilities operating on containers inheriting from MetaInfoInterface |
| CMetaInfoRegistry | Registry which assigns unique integer indices to strings |
| CMetaInfoVisualizer | MetaInfoVisualizer is a visualizer class for all classes that use one MetaInfo object as member |
| CMinimumDistance | Basic data structure for distances between clusters |
| CModelDescription | Stores the name and parameters of a model |
| CModification | Meta information about chemical modification of a sample |
| CModificationDefinition | Representation of modification definition |
| CModificationDefinitionsSet | Representation of a set of modification definitions |
| CModificationsDB | Database which holds all residue modifications from UniMod |
| CModificationVisualizer | Class that displays all meta information of modification objects |
| CModifiedPeptideGenerator | |
| CModifierRep | Implements modification for suffix arrays |
| CMorphologicalFilter | This class implements baseline filtering operations using methods from mathematical morphology |
| CMRMAssay | Generate assays from a TargetedExperiment |
| CMRMDecoy | This class generates a TargetedExperiment object with decoys based on a TargetedExperiment object |
| CMRMFeature | A multi-chromatogram MRM feature |
| CMRMFeatureFinderScoring | The MRMFeatureFinder finds and scores peaks of transitions that co-elute |
| CMRMFeatureOpenMS | An implementation of the OpenSWATH MRM Feature Access interface using OpenMS |
| CMRMFragmentSelection | This class can select appropriate fragment ions of an MS/MS spectrum of a peptide |
| CMRMIonSeries | Generate theoretical fragment ion series for use in MRMAssay and MRMDecoy |
| CMRMRTNormalizer | The MRMRTNormalizer will find retention time peptides in data |
| CMRMTransitionGroup | The representation of a group of transitions in a targeted proteomics experiment |
| CMRMTransitionGroupPicker | The MRMTransitionGroupPicker finds peaks in chromatograms that belong to the same precursors |
| ►CMS1FeatureMerger | |
| COPERATOR_FEATURE_TR | |
| CMS1Signal | |
| CMS2ConsensusSpectrum | |
| CMS2Feature | |
| CMS2File | MS2 input file adapter |
| CMS2Fragment | |
| CMS2Info | |
| ►CMSChromatogram | The representation of a chromatogram |
| CMZLess | Comparator for the retention time |
| CMSDataAggregatingConsumer | Aggregates spectra by retention time |
| CMSDataCachedConsumer | Transforming and cached writing consumer of MS data |
| CMSDataChainingConsumer | Consumer class that passes all consumed data through a set of operations |
| CMSDataSqlConsumer | A data consumer that inserts data into a SQL database |
| CMSDataStoringConsumer | Consumer class that simply stores the data |
| CMSDataTransformingConsumer | Transforming consumer of MS data |
| CMSDataWritingConsumer | Consumer class that writes MS data to disk using the mzML format |
| ►CMSExperiment | In-Memory representation of a mass spectrometry experiment |
| CContainerAdd_ | Helper class to add either general data points in set2DData or use mass traces from meta values |
| CContainerAdd_< ContainerValueType, false > | |
| CContainerAdd_< ContainerValueType, true > | |
| CMsInspectFile | File adapter for MsInspect files |
| ►CMSNumpressCoder | Class to encode and decode data encoded with MSNumpress |
| CNumpressConfig | Configuration class for MSNumpress |
| CMSPeak | |
| CMSPFile | File adapter for MSP files (NIST spectra library) |
| ►CMSQuantifications | |
| CAnalysisSummary | |
| CAssay | |
| CMSSim | Central class for simulation of mass spectrometry experiments |
| ►CMSSpectrum | The representation of a 1D spectrum |
| CRTLess | Comparator for the retention time |
| CMultiGradient | A gradient of multiple colors and arbitrary distances between colors |
| CMultiGradientSelector | A widget witch allows constructing gradients of multiple colors |
| ►CMultiplexClustering | Clusters results from multiplex filtering |
| CMultiplexDistance | Scaled Euclidean distance for clustering |
| ►CMultiplexDeltaMasses | Data structure for mass shift pattern |
| CDeltaMass | Mass shift with corresponding label set |
| ►CMultiplexDeltaMassesGenerator | Generates complete list of all possible mass shifts due to isotopic labelling |
| CLabel | Complete label information |
| ►CMultiplexFiltering | Base class for filtering centroided and profile data for peak patterns |
| CBlackListEntry | Structure for peak blacklisting |
| CPeakReference | Structure for peak position in neighbouring spectra |
| CMultiplexFilteringCentroided | Filters centroided data for peak patterns |
| CMultiplexFilteringProfile | Filters centroided and profile data for peak patterns |
| CMultiplexFilterResult | Data structure storing all peaks (and optionally their raw data points) corresponding to one specific peak pattern |
| CMultiplexFilterResultPeak | Data structure storing a single peak that passed all filters |
| CMultiplexFilterResultRaw | Data structure storing a single raw data point that passed all filters |
| CMultiplexIsotopicPeakPattern | Data structure for pattern of isotopic peaks |
| CMzDataFile | File adapter for MzData files |
| CMzIdentMLFile | File adapter for MzIdentML files |
| CMzMLFile | File adapter for MzML files |
| CMzMLSpectrumDecoder | A class to decode input strings that contain an mzML chromatogram or spectrum tag |
| CMzMLSwathFileConsumer | On-disk mzML implementation of FullSwathFileConsumer |
| CMzQuantMLFile | File adapter for MzQuantML files |
| CMzTab | Data model of MzTab files. Please see the official MzTab specification at https://code.google.com/p/mztab/ |
| CMzTabAssayMetaData | |
| CMzTabBoolean | |
| CMzTabContactMetaData | |
| CMzTabCVMetaData | |
| CMzTabDouble | |
| CMzTabDoubleList | |
| CMzTabFile | File adapter for MzTab files |
| CMzTabInstrumentMetaData | |
| CMzTabInteger | |
| CMzTabIntegerList | |
| CMzTabMetaData | |
| CMzTabModification | |
| CMzTabModificationList | |
| CMzTabModificationMetaData | |
| CMzTabMSRunMetaData | |
| CMzTabNullAbleBase | |
| CMzTabNullAbleInterface | |
| CMzTabNullNaNAndInfAbleBase | |
| CMzTabNullNaNAndInfAbleInterface | |
| CMzTabParameter | |
| CMzTabParameterList | |
| CMzTabPeptideSectionRow | |
| CMzTabProteinSectionRow | |
| CMzTabPSMSectionRow | |
| CMzTabSampleMetaData | |
| CMzTabSmallMoleculeSectionRow | |
| CMzTabSoftwareMetaData | |
| CMzTabSpectraRef | |
| CMzTabString | |
| CMzTabStringList | |
| CMzTabStudyVariableMetaData | |
| ►CMZTrafoModel | Create and apply models of a mass recalibration function |
| CRTLess | Comparator by position. As this class has dimension 1, this is basically an alias for MZLess |
| CMzXMLFile | File adapter for MzXML 3.1 files |
| CNetworkGetRequest | |
| CNeutralLossDiffFilter | NeutralLossDiffFilter returns the total intensity ob peak pairs whose m/z difference can be explained by a neutral loss |
| CNeutralLossMarker | NeutralLossMarker marks peak pairs which could represent an ion an its neutral loss (water, ammonia) |
| CNLargest | NLargest removes all but the n largest peaks |
| CNonNegativeLeastSquaresSolver | Wrapper for a non-negative least squares (NNLS) solver |
| CNoopMSDataConsumer | Consumer class that performs no operation |
| CNoopMSDataWritingConsumer | Consumer class that perform no operation |
| CNormalizer | Normalizes the peak intensities spectrum-wise |
| CO18Labeler | Simulate O-18 experiments |
| COfflinePrecursorIonSelection | Implements different algorithms for precursor ion selection |
| COMSSACSVFile | File adapter for OMSSACSV files |
| COMSSAXMLFile | Used to load OMSSAXML files |
| COnDiscMSExperiment | Representation of a mass spectrometry experiment on disk |
| COpenSwath_Scores | A structure to hold the different scores computed by OpenSWATH |
| COpenSwath_Scores_Usage | A structure to store which scores should be used by the Algorithm |
| COpenSwathDataAccessHelper | Several helpers to convert OpenMS datastructures to structures that implement the OpenSWATH interfaces |
| COpenSwathHelper | A helper class that is used by several OpenSWATH tools |
| COpenSwathOSWWriter | Class to write out an OpenSwath OSW SQLite output (PyProphet input) |
| COpenSwathRetentionTimeNormalization | Simple OpenSwathWorkflow to perform RT and m/z correction based on a set of known peptides |
| COpenSwathScoring | A class that calls the scoring routines |
| COpenSwathTSVWriter | Class to write out an OpenSwath TSV output (mProphet input) |
| COpenSwathWorkflow | Class to execute an OpenSwath Workflow |
| COpenSwathWorkflowSonar | Class to execute an OpenSwath Workflow for SONAR data |
| ►COptimizePeakDeconvolution | This class provides the deconvolution of peak regions using non-linear optimization |
| CData | Class containing the data needed for optimization |
| ►COptimizePick | This class provides the non-linear optimization of the peak parameters |
| CData | |
| COptPeakFunctor | |
| ►COPXLDataStructs | |
| CAASeqWithMass | The AASeqWithMass struct represents a normal peptide with its precomputed mass |
| CAASeqWithMassComparator | The AASeqWithMassComparator is a comparator for AASeqWithMass objects |
| CCrossLinkSpectrumMatch | The CrossLinkSpectrumMatch struct represents a PSM between a ProteinProteinCrossLink and a spectrum in OpenPepXL |
| CPreprocessedPairSpectra | The PreprocessedPairSpectra struct represents the result of comparing a light and a heavy labeled spectra to each other |
| CProteinProteinCrossLink | The ProteinProteinCrossLink struct represents a cross-link between two peptides in OpenPepXL |
| CXLPrecursor | The XLPrecursor struct represents a cross-link candidate in the process of filtering candidates by precursor masses in OpenPepXL |
| CXLPrecursorComparator | The XLPrecursorComparator is a comparator for XLPrecursors, that allows direct comparison of the XLPrecursor precursor mass with double numbers |
| COPXLHelper | Functions needed by OpenPepXL and OpenPepXLLF to reduce duplicated code |
| COPXLSpectrumProcessingAlgorithms | |
| CPairComparatorFirstElement | Class for comparison of std::pair using first ONLY e.g. for use with std::sort |
| CPairComparatorFirstElementMore | Class for comparison of std::pair using first ONLY e.g. for use with std::sort |
| CPairComparatorSecondElement | Class for comparison of std::pair using second ONLY e.g. for use with std::sort |
| CPairComparatorSecondElementMore | Class for comparison of std::pair using second ONLY e.g. for use with std::sort |
| CPairMatcherFirstElement | Class for comparison of std::pair using first ONLY e.g. for use with std::sort |
| CPairMatcherSecondElement | Struct for comparison of std::pair using second ONLY e.g. for use with std::sort |
| ►CParam | Management and storage of parameters / INI files |
| CParamEntry | Parameter entry used to store the actual information inside of a Param entry |
| ►CParamIterator | Forward const iterator for the Param class |
| CTraceInfo | Struct that captures information on entered / left nodes for ParamIterator |
| CParamNode | Node inside a Param object which is used to build the internal tree |
| CParamEditor | A GUI for editing or viewing a Param object |
| CParameterInformation | Struct that captures all information of a command line parameter |
| CParamXMLFile | The file pendant of the Param class used to load and store the param datastructure as paramXML |
| CParentPeakMower | ParentPeakMower gets rid of high peaks that could stem from unfragmented precursor ions |
| ►CPeak1D | A 1-dimensional raw data point or peak |
| CIntensityLess | |
| CMZLess | Comparator by m/z position |
| CPositionLess | Comparator by position. As this class has dimension 1, this is basically an alias for MZLess |
| ►CPeak2D | A 2-dimensional raw data point or peak |
| CIntensityLess | |
| CMZLess | Comparator by m/z position |
| CPositionLess | Comparator by position. Lexicographical comparison (first RT then m/z) is done |
| CRTLess | Comparator by RT position |
| CPeakAlignment | Make a PeakAlignment of two PeakSpectra |
| CPeakCandidate | A small structure to hold peak candidates |
| CPeakFileOptions | Options for loading files containing peak data |
| CPeakIndex | Index of a peak or feature |
| CPeakIntensityPredictor | Predict peak heights of peptides based on Local Linear Map model |
| CPeakMarker | PeakMarker marks peaks that seem to fulfill some criterion |
| ►CPeakPickerCWT | This class implements a peak picking algorithm using wavelet techniques |
| CPeakArea_ | Class for the internal peak representation |
| ►CPeakPickerHiRes | This class implements a fast peak-picking algorithm best suited for high resolution MS data (FT-ICR-MS, Orbitrap). In high resolution data, the signals of ions with similar mass-to-charge ratios (m/z) exhibit little or no overlapping and therefore allow for a clear separation. Furthermore, ion signals tend to show well-defined peak shapes with narrow peak width |
| CPeakBoundary | Structure for peak boundaries |
| CPeakPickerIterative | This class implements a peak-picking algorithm for high-resolution MS data (specifically designed for TOF-MS data) |
| ►CPeakPickerMaxima | This class implements a fast peak-picking algorithm best suited for high resolution MS data (FT-ICR-MS, Orbitrap). In high resolution data, the signals of ions with similar mass-to-charge ratios (m/z) exhibit little or no overlapping and therefore allow for a clear separation. Furthermore, ion signals tend to show well-defined peak shapes with narrow peak width |
| CPeakCandidate | The PeakCandidate describes the output of the peak picker |
| CPeakPickerMRM | The PeakPickerMRM finds peaks a single chromatogram |
| CPeakPickerSH | |
| ►CPeakShape | Internal representation of a peak shape (used by the PeakPickerCWT) |
| CPositionLess | Comparison of mz_positions |
| CPeakSpectrumCompareFunctor | Base class for compare functors of spectra, that return a similarity value for two spectra |
| CPeakTypeEstimator | Estimates if the data of a spectrum is raw data or peak data |
| CPeakWidthEstimator | Rough estimation of the peak width at m/z |
| CPepIterator | Abstract base class for different peptide iterators |
| CPepNovoInfile | PepNovo input file adapter |
| CPepNovoOutfile | Representation of a PepNovo output file |
| ►CPeptideAndProteinQuant | Helper class for peptide and protein quantification based on feature data annotated with IDs |
| CPeptideData | Quantitative and associated data for a peptide |
| CProteinData | Quantitative and associated data for a protein |
| CStatistics | Statistics for processing summary |
| CPeptideEvidence | Representation of a peptide evidence |
| ►CPeptideHit | Representation of a peptide hit |
| CPeakAnnotation | Contains annotations of a peak |
| CPepXMLAnalysisResult | Analysis Result (containing search engine / prophet results) |
| CRankLess | Lesser predicate for scores of hits |
| CScoreLess | Lesser predicate for scores of hits |
| CScoreMore | Greater predicate for scores of hits |
| CSequenceLessComparator | Lesser predicate for (modified) sequence of hits |
| CPeptideHitVisualizer | Class that displays all meta information for PeptideHit objects |
| CPeptideIdentification | Represents the peptide hits for a spectrum |
| CPeptideIdentificationVisualizer | Class that displays all meta information for PeptideIdentification objects |
| CPeptideIndexing | Refreshes the protein references for all peptide hits in a vector of PeptideIdentifications and adds target/decoy information |
| CPeptideProteinResolution | Resolves shared peptides based on protein scores |
| ►CPepXMLFile | Used to load and store PepXML files |
| CAminoAcidModification | |
| CPepXMLFileMascot | Used to load Mascot PepXML files |
| ►CPercolatorFeatureSetHelper | Percolator feature set and integration helper |
| Clq_PeptideEvidence | For accession dependent sorting of PeptideEvidences |
| Clq_ProteinHit | For accession dependent sorting of ProteinHits |
| CPercolatorOutfile | Class for reading Percolator tab-delimited output files |
| CPlainMSDataWritingConsumer | Consumer class that writes MS data to disk using the mzML format |
| CPointerComparator | Wrapper that takes a comparator for `something' and makes a comparator for pointers to `something' out of it. Normally you should use the make-function pointerComparator() because then you do not need to specify the template arguments |
| CPoseClusteringAffineSuperimposer | A superimposer that uses a voting scheme, also known as pose clustering, to find a good affine transformation |
| CPoseClusteringShiftSuperimposer | A superimposer that uses a voting scheme, also known as pose clustering, to find a good shift transformation |
| CPrecisionWrapper | Wrapper class to implement output with appropriate precision. See precisionWrapper() |
| CPrecursor | Precursor meta information |
| ►CPrecursorIonSelection | This class implements different precursor ion selection strategies |
| CSeqTotalScoreMore | Compare by score |
| CTotalScoreMore | Compare by score |
| CPrecursorIonSelectionPreprocessing | This class implements the database preprocessing needing for precursor ion selection |
| CPrecursorMassComparator | |
| CPrecursorVisualizer | Class that displays all meta information for Precursor objects |
| CProbablePhosphoSites | |
| CProcessData | |
| CProduct | Product meta information |
| CProductModel | Class for product models i.e. models with D independent dimensions |
| CProductModel< 2 > | The class template is only implemented for D=2 because we use Peak2D here |
| CProductVisualizer | Class that displays all meta information for Product objects |
| ►CProgressLogger | Base class for all classes that want to report their progress |
| CProgressLoggerImpl | This class represents an actual implementation of a logger |
| ►CProteinHit | Representation of a protein hit |
| CScoreLess | Lesser predicate for scores of hits |
| CScoreMore | Greater predicate for scores of hits |
| CProteinHitVisualizer | Class that displays all meta information for ProteinHit objects |
| ►CProteinIdentification | Representation of a protein identification run |
| CProteinGroup | Bundles multiple (e.g. indistinguishable) proteins in a group |
| CSearchParameters | Search parameters of the DB search |
| CProteinIdentificationVisualizer | Class that displays all meta information for ProteinIdentification objects |
| CProteinInference | [experimental class] given a peptide quantitation, infer corresponding protein quantities |
| ►CProteinResolver | Helper class for peptide and protein quantification based on feature data annotated with IDs |
| CISDGroup | |
| CMSDGroup | Representation of an msd group. Contains peptides, proteins and a pointer to its ISD group |
| CPeptideEntry | Peptide. First in silico. If experimental is set to true it is MS/MS derived |
| CProteinEntry | Protein from FASTA file |
| CResolverResult | |
| CProtonDistributionModel | A proton distribution model to calculate the proton distribution over charged peptides |
| CProtXMLFile | Used to load (storing not supported, yet) ProtXML files |
| CPScore | Implementation of the PScore PSM scoring algorithm |
| ►CPSLPFormulation | Implements ILP formulation of precursor selection problems |
| CIndexLess | |
| CIndexTriple | Struct that holds the indices of the precursors in the feature map and the ilp formulation |
| CScanLess | |
| CVariableIndexLess | |
| CPSProteinInference | This class implements protein inference for the precursor ion selection strategies |
| CPTMXMLFile | Used to load and store PTMXML files |
| CQApplicationTOPP | Extension to the QApplication for running TOPPs GUI tools |
| ►CQcMLFile | File adapter for QcML files |
| CAttachment | Representation of an attachment |
| CQualityParameter | Representation of a quality parameter |
| CQTCluster | A representation of a QT cluster used for feature grouping |
| CQTClusterFinder | A variant of QT clustering for the detection of feature groups |
| CQuantitativeExperimentalDesign | Merge files according to experimental design |
| CRangeManager | Handles the management of a position and intensity range |
| CRawData | |
| ►CRawMSSignalSimulation | Simulates MS signals for a given set of peptides |
| CContaminantInfo | |
| CRawTandemMSSignalSimulation | Simulates tandem MS signals for a given set of peptides |
| ►CReactionMonitoringTransition | This class stores a SRM/MRM transition |
| CProductMZLess | Comparator by Product ion MZ |
| CRegularSwathFileConsumer | In-memory implementation of FullSwathFileConsumer |
| CResidue | Representation of a residue |
| CResidueDB | Residue data base which holds residues |
| CResidueModification | Representation of a modification |
| CReverseComparator | Wrapper that reverses (exchanges) the two arguments of a comparator. Normally you should use the make-function reverseComparator() because then you do not need to specify the template arguments |
| CRichPeak2D | A 2-dimensional raw data point or peak with meta information |
| CRNPxlMarkerIonExtractor | |
| CRNPxlModificationMassesResult | |
| CRNPxlModificationsGenerator | |
| CRNPxlReport | |
| CRNPxlReportRow | |
| CRNPxlReportRowHeader | |
| CRTSimulation | Simulates/Predicts retention times for peptides or peptide separation |
| CRWrapper | R-Wrapper Class |
| CSample | Meta information about the sample |
| CSampleTreatment | Base class for sample treatments (Digestion, Modification, Tagging, ...) |
| CSampleVisualizer | Class that displays all meta information of sample objects |
| CSaveImageDialog | Dialog for saving an image |
| CSavitzkyGolayFilter | Computes the Savitzky-Golay filter coefficients using QR decomposition |
| CScaler | Scaler scales the peak by ranking the peaks and assigning intensity according to rank |
| CScanWindow | Scan window description |
| CScanWindowVisualizer | Class that displays all meta information for ScanWindow objects |
| CSeedListGenerator | Generate seed lists for feature detection |
| CSequestInfile | Sequest input file adapter |
| CSequestOutfile | Representation of a Sequest output file |
| CSHFeature | |
| ►CSignalToNoiseEstimator | This class represents the abstract base class of a signal to noise estimator |
| CGaussianEstimate | Protected struct to store parameters my, sigma for a Gaussian distribution |
| CSignalToNoiseEstimatorMeanIterative | Estimates the signal/noise (S/N) ratio of each data point in a scan based on an iterative scheme which discards high intensities |
| CSignalToNoiseEstimatorMedian | Estimates the signal/noise (S/N) ratio of each data point in a scan by using the median (histogram based) |
| ►CSignalToNoiseEstimatorMedianRapid | Estimates the signal/noise (S/N) ratio of each data point in a scan by using the median (window based) |
| CNoiseEstimator | Class to compute the noise value at a given position |
| CSignalToNoiseOpenMS | An implementation of the OpenSWATH SignalToNoise Access interface using OpenMS |
| CSILACLabeler | Simulate SILAC experiments |
| CSimpleOpenMSSpectraFactory | A factory method that returns two ISpectrumAccess implementations |
| CSimplePairFinder | This class implements a simple point pair finding algorithm |
| ►CSimpleSVM | Simple interface to support vector machines for classification (via LIBSVM) |
| CPrediction | SVM prediction result |
| CSingleLinkage | SingleLinkage ClusterMethod |
| CSingletonRegistry | Holds pointers to unique instance of a singleton factory |
| CSiriusMSFile | |
| ►CSiriusMzTabWriter | |
| CSiriusAdapterHit | Internal structure used in SiriusAdapter that is used for the conversion of the sirius output to an mzTab |
| CSiriusAdapterIdentification | |
| CSiriusAdapterRun | |
| CSoftware | Description of the software used for processing |
| CSoftwareVisualizer | Class that displays all meta information for Software objects |
| CSONARScoring | Scoring of an spectrum using SONAR data |
| CSourceFile | Description of a file location, used to store the origin of (meta) data |
| CSourceFileVisualizer | Class that displays all meta information for SourceFile objects |
| ►CSparseVector | SparseVector implementation. The container will not actually store a specified type of element - the sparse element, e.g. zero (by default) |
| CSparseVectorConstIterator | Const_iterator for SparseVector |
| CSparseVectorConstReverseIterator | Const_reverse_iterator for SparseVector |
| CSparseVectorIterator | Random access iterator for SparseVector including the hop() function to jump to the next non-sparse element |
| CSparseVectorReverseIterator | Random access reverse iterator for SparseVector including the hop() function to jump to the next non-sparse element |
| CValueProxy | Class ValueProxy allows the SparseVector to differentiate between writing and reading, so zeros can be ignored See "more effective c++" section 30 |
| CSpecArrayFile | File adapter for SpecArray (.pepList) files |
| CSpectraIdentificationViewWidget | Tabular visualization / selection of identified spectra |
| CSpectralMatch | |
| CSpectralMatchScoreComparator | |
| ►CSpectraMerger | Merges blocks of MS or MS2 spectra |
| CSpectraDistance_ | |
| CSpectraSTSimilarityScore | Similarity score of SpectraST |
| CSpectraViewWidget | Hierarchical visualization and selection of spectra |
| CSpectrum1DCanvas | Canvas for visualization of one or several spectra |
| CSpectrum1DGoToDialog | Simple goto/set visible area dialog for exact placement of the viewing window |
| CSpectrum1DWidget | Widget for visualization of several spectra |
| CSpectrum2DCanvas | Canvas for 2D-visualization of peak map, feature map and consensus map data |
| CSpectrum2DGoToDialog | GoTo dialog used to zoom to a m/z and retention time range or to a feature |
| CSpectrum2DWidget | Widget for 2D-visualization of peak map and feature map data |
| CSpectrum3DCanvas | Canvas for 3D-visualization of peak map data |
| CSpectrum3DOpenGLCanvas | OpenGL Canvas for 3D-visualization of map data |
| CSpectrum3DWidget | Widget for 3D-visualization of map data |
| CSpectrumAccessOpenMS | An implementation of the OpenSWATH Spectrum Access interface using OpenMS |
| CSpectrumAccessOpenMSCached | An implementation of the Spectrum Access interface using on-disk caching |
| CSpectrumAccessOpenMSInMemory | An implementation of the OpenSWATH Spectrum Access interface completely in memory |
| CSpectrumAccessQuadMZTransforming | A transforming m/z wrapper around spectrum access using a quadratic equation |
| CSpectrumAccessTransforming | An abstract base class implementing a transforming wrapper around spectrum access |
| CSpectrumAddition | The SpectrumAddition is used to add up individual spectra |
| CSpectrumAlignment | Aligns the peaks of two sorted spectra Method 1: Using a banded (width via 'tolerance' parameter) alignment if absolute tolerances are given. Scoring function is the m/z distance between peaks. Intensity does not play a role! |
| CSpectrumAlignmentDialog | Lets the user select two spectra and set the parameters for the spectrum alignment |
| CSpectrumAlignmentScore | Similarity score via spectra alignment |
| CSpectrumAnnotator | Annotates spectra from identifications and theoretical spectra or identifications from spectra and theoretical spectra matching with various options |
| CSpectrumCanvas | Base class for visualization canvas classes |
| CSpectrumCheapDPCorr | SpectrumCheapDPCorr calculates an optimal alignment on stick spectra |
| CSpectrumIdentification | Represents a object which can store the information of an analysisXML instance |
| CSpectrumLookup | Helper class for looking up spectra based on different attributes |
| ►CSpectrumMetaDataLookup | Helper class for looking up spectrum meta data |
| CSpectrumMetaData | Meta data of a spectrum |
| CSpectrumPrecursorComparator | SpectrumPrecursorComparator compares just the parent mass of two spectra |
| CSpectrumSettings | Representation of 1D spectrum settings |
| CSpectrumSettingsVisualizer | Class that displays all meta information for SpectrumSettings objects |
| CSpectrumWidget | Base class for spectrum widgets |
| CSplinePackage | Fundamental data structure for SplineSpectrum |
| ►CSplineSpectrum | Data structure for spline interpolation of MS1 spectra |
| CNavigator | Iterator class for access of spline packages |
| CSqMassFile | An class that uses on-disk SQLite database to read and write spectra and chromatograms |
| CSqrtMower | Scales the intensity of peaks to the sqrt |
| CStablePairFinder | This class implements a pair finding algorithm for consensus features |
| CSteinScottImproveScore | Similarity score based of Stein & Scott |
| CStopWatch | StopWatch Class |
| CStreamHandler | Provides a central class to register globally used output streams. Currently supported streams are |
| CString | A more convenient string class |
| CStringListUtils | Utilities operating on lists of Strings |
| ►CStringUtils | |
| Creal_policies_NANfixed_ | |
| CStringView | StringView provides a non-owning view on an existing string |
| CSummary | Summary of fitting results |
| CSuperHirnParameters | SuperHirn parameters singleton class containing all static configuration variables |
| CSuperHirnUtil | |
| CSVMData | Data structure used in SVMWrapper |
| ►CSvmTheoreticalSpectrumGenerator | Simulates MS2 spectra with support vector machines |
| CDescriptorSet | A set of descriptors for a single training row |
| CIonType | Nested class |
| CSvmModelParameterSet | Simple container storing the model parameters required for simulation |
| CSvmTheoreticalSpectrumGeneratorSet | Loads SvmTheoreticalSpectrumGenerator instances for different charges |
| CSvmTheoreticalSpectrumGeneratorTrainer | Train SVM models that are used by SvmTheoreticalSpectrumGenerator |
| CSVMWrapper | Serves as a wrapper for the libsvm |
| CSVOutStream | Stream class for writing to comma/tab/...-separated values files |
| CSwathFile | File adapter for Swath files |
| CSwathMapMassCorrection | A class containing correction functions for Swath MS maps |
| CSwathWindowLoader | Class to read a file describing the Swath Windows |
| ►CSysInfo | Some functions to get system information |
| CMemUsage | A convenience class to report either absolute or delta (between two timepoints) RAM usage |
| CTagging | Meta information about tagging of a sample e.g. ICAT labeling |
| CTaggingVisualizer | Class that displays all meta information of tagging objects |
| CTargetedExperiment | A description of a targeted experiment containing precursor and production ions |
| CTextFile | This class provides some basic file handling methods for text files |
| CTheoreticalSpectrumGenerationDialog | Dialog which allows to enter an AA sequence and generates a theoretical spectrum for it |
| CTheoreticalSpectrumGenerator | Generates theoretical spectra with various options |
| CTheoreticalSpectrumGeneratorXLMS | Generates theoretical spectra for cross-linked peptides |
| CThresholdMower | ThresholdMower removes all peaks below a threshold |
| CTICFilter | TICFilter calculates TIC |
| CTMTElevenPlexQuantitationMethod | TMT 11plex quantitation to be used with the IsobaricQuantitation |
| CTMTSixPlexQuantitationMethod | TMT 6plex quantitation to be used with the IsobaricQuantitation |
| CTMTTenPlexQuantitationMethod | TMT 10plex quantitation to be used with the IsobaricQuantitation |
| CTOFCalibration | This class implements an external calibration for TOF data using external calibrant spectra |
| CToolDescriptionFile | File adapter for ToolDescriptor files |
| CToolHandler | |
| CToolsDialog | TOPP tool selection dialog |
| CTOPPASBase | Main window of the TOPPAS tool |
| CTOPPASEdge | An edge representing a data flow in TOPPAS |
| CTOPPASInputFileDialog | Dialog which allows to specify an input file |
| CTOPPASInputFileListVertex | A vertex representing an input file list |
| CTOPPASInputFilesDialog | Dialog which allows to specify a list of input files |
| CTOPPASIOMappingDialog | Dialog which allows to configure the input/output parameter mapping of an edge |
| CTOPPASLogWindow | QTextEdit implementation with a "clear" button in the context menu |
| CTOPPASMergerVertex | A special vertex that allows to merge several inputs |
| CTOPPASOutputFileListVertex | A vertex representing an output file list |
| CTOPPASOutputFilesDialog | Dialog which allows to specify the directory for the output files |
| CTOPPASResource | Represents a data resource for TOPPAS workflows |
| CTOPPASResources | A dictionary mapping string keys to lists of TOPPASResource objects |
| ►CTOPPASScene | A container for all visual items of a TOPPAS workflow |
| CTOPPProcess | Stores the information for a TOPP process |
| CTOPPASSplitterVertex | A special vertex that allows to split a list of inputs |
| CTOPPASTabBar | Convenience tab bar implementation |
| CTOPPASToolConfigDialog | TOPP tool configuration dialog |
| ►CTOPPASToolVertex | A vertex representing a TOPP tool |
| CIOInfo | Stores the information for input/output files/lists |
| CTOPPASTreeView | Tree view implementation for the list of TOPP tools |
| ►CTOPPASVertex | The base class of the different vertex classes |
| CTOPPASFilenames | |
| CVertexRoundPackage | Info for one edge and round, to be passed to next node |
| CTOPPASVertexNameDialog | Dialog which allows to change the name of an input/output vertex |
| CTOPPASWidget | Widget visualizing and allowing to edit TOPP pipelines |
| CTOPPBase | Base class for TOPP applications |
| CTOPPViewBase | Main window of TOPPView tool |
| CTOPPViewIdentificationViewBehavior | Behavior of TOPPView in identification mode |
| CTOPPViewOpenDialog | Dataset opening options for TOPPView |
| CTOPPViewSpectraViewBehavior | Behavior of TOPPView in spectra view mode |
| ►CTraceFitter | Abstract fitter for RT profile fitting |
| CGenericFunctor | |
| CModelData | |
| CTraMLFile | File adapter for HUPO PSI TraML files |
| CTransformationDescription | Generic description of a coordinate transformation |
| CTransformationModel | Base class for transformation models |
| CTransformationModelBSpline | B-spline (non-linear) model for transformations |
| ►CTransformationModelInterpolated | Interpolation model for transformations |
| CInterpolator | The class defines a generic interpolation technique used in the TransformationModelInterpolated |
| CTransformationModelLinear | Linear model for transformations |
| CTransformationModelLowess | Lowess (non-linear) model for transformations |
| CTransformationXMLFile | Used to load and store TransformationXML files |
| CTransitionGroupOpenMS | An implementation of the OpenSWATH Transition Group Access interface using OpenMS |
| CTransitionPQPReader | This class can convert TraML and PQP files into each other |
| ►CTransitionTSVReader | This class can convert TraML and TSV files into each other |
| CTSVTransition | Internal structure to represent a transition |
| CTrypticIterator | Finds all tryptic Peptides with every missed cleavage |
| ►CTwoDOptimization | This class provides the two-dimensional optimization of the picked peak parameters |
| CData | Helper struct (contains the size of an area and a raw data container) |
| CTwoDOptFunctor | |
| CUnaryComposeFunctionAdapter | Represents the function object unary adapter |
| CUnimodXMLFile | Used to load XML files from unimod.org files |
| CUniqueIdGenerator | A generator for unique ids |
| CUniqueIdIndexer | A base class for random access containers for classes derived from UniqueIdInterface that adds functionality to convert a unique id into an index into the container |
| CUniqueIdInterface | A base class defining a common interface for all classes having a unique id |
| CUnnormalizedComparator | Exception thrown if clustering is attempted without a normalized compare functor |
| CUpdateCheck | Helper Functions to perform an update query to the OpenMS REST server |
| ►CVersionInfo | Version information class |
| CVersionDetails | |
| CWeightWrapper | Encapsulated weight queries to simplify mono vs average weight computation |
| CWindowMower | WindowMower augments the highest peaks in a sliding or jumping window |
| CXMassFile | File adapter for 'XMass Analysis (fid)' files |
| CXMLValidator | Validator for XML files |
| CXQuestResultXMLFile | |
| CXQuestScores | An implementation of the scores for cross-link identification from the xQuest algorithm (O. Rinner et al., 2008, "Identification of cross-linked peptides from large sequence databases") |
| CXTandemInfile | XTandem input file |
| CXTandemXMLFile | Used to load XTandemXML files |
| CZhangSimilarityScore | Similarity score of Zhang |
| CZlibCompression | Compresses and uncompresses data using zlib |
| ►NOpenSwath | |
| CCSVWriter | |
| CDataMatrix | |
| CIDataFrameWriter | |
| CIFeature | |
| CIMRMFeature | |
| CISignalToNoise | |
| CISpectrumAccess | The interface of a mass spectrometry experiment |
| CITransitionGroup | |
| CLightCompound | |
| CLightModification | |
| CLightProtein | |
| CLightTargetedExperiment | |
| CLightTransition | |
| Cmean_and_stddev | Functor to compute the mean and stddev of sequence using the std::foreach algorithm |
| CMockFeature | Mock object implementing IFeature |
| CMockMRMFeature | Mock object implementing IMRMFeature |
| CMockSignalToNoise | Mock object implementing ISignalToNoise |
| CMockTransitionGroup | Mock object implementing ITransitionGroup |
| CMRMScoring | This class implements different scores for peaks found in SRM/MRM |
| CmySqrt | |
| COSBinaryDataArray | The datastructures used by the OpenSwath interfaces |
| COSChromatogram | A single chromatogram |
| COSChromatogramMeta | Identifying information for a chromatogram |
| COSSpectrum | The structure that captures the generation of a peak list (including the underlying acquisitions) |
| ►COSSpectrumMeta | Identifying information for a spectrum |
| CRTLess | Comparator for the retention time |
| CPeptide | |
| CProtein | |
| CSwathMap | Data structure to hold one SWATH map with information about upper / lower isolation window and whether the map is MS1 or MS2 |
| CTargetedExperiment | |
| CTransitionHelper | |
| ►Nseqan | |
| CAdaptedIdentity | |
| CPAM30MS | |
| CScoringMatrixData_< int, AminoAcid, AdaptedIdentity > | |
| CScoringMatrixData_< int, AminoAcid, PAM30MS > | |
| CDeisotoper | |
| CFragmentAnnotationDetail_ | Single fragment annotation |
| CFragmentAnnotationHelper | |
| Cglp_prob | |
| CMetaProSIPClustering | |
| CMetaProSIPDecomposition | |
| CMetaProSIPInterpolation | |
| CMetaProSIPReporting | |
| CMetaProSIPXICExtraction | |
| CQApplication | |
| CQDate | |
| CQDateTime | |
| CQDialog | |
| CQFileSystemWatcher | |
| CQGLWidget | |
| CQGraphicsItem | |
| CQGraphicsScene | |
| CQGraphicsView | |
| CQItemDelegate | |
| CQLineEdit | |
| CQListWidget | |
| CQMainWindow | |
| CQObject | |
| CQProcess | |
| CQTabBar | |
| CQTextEdit | |
| CQTreeWidget | |
| CQWidget | |
| CQWorkspace | |
| CRateScorePair | |
| CRIALess | |
| CRIntegration | |
| ►CRNPxlSearch | |
| CAnnotatedHit | Slimmer structure as storing all scored candidates in PeptideHit objects takes too much space |
| CFragmentAdductDefinition_ | |
| CSequenceLess | |
| CSimpleSearchEngine | |
| CSIPIncorporation | Datastructure for reporting an incorporation event |
| CSIPPeptide | Datastructure for reporting a peptide with one or more incorporation rates comparator for vectors of SIPPeptides based on their size. Used to sort by group size |
| CSizeLess | |
| CTOPPMetaProSIP | |
| CTOPPOpenPepXL | |
| CTOPPOpenPepXLLF | |
| CTOPPRNPxl |