 |
OpenMS
|
 |
OpenMS
|
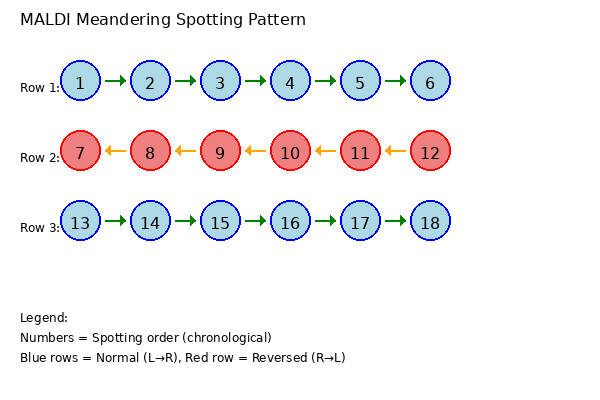
Repairs MALDI experiments which were spotted line by line in a meandering pattern.
| pot pot pot |
|---|
|
Problem Description:
MALDI (Matrix-Assisted Laser Desorption/Ionization) spotting robots often apply samples to target plates in a "meandering" or "snake-like" pattern to optimize spotting efficiency. This means:
When the mass spectrometer reads these spots in physical order (left to right for all rows), the spectra from alternating rows have incorrect retention time assignments, appearing in reverse chronological order compared to when they were spotted.
Solution:
DeMeanderize corrects this by:
Algorithm Details:
The tool processes spectra sequentially and assigns pseudo-RT values:
The algorithm maintains:
After processing num_spots_per_row MS1 spectra, the algorithm switches to the next row and toggles the reversal flag.
Parameters:
num_spots_per_row: Number of spots in each row of the plate (default: 48)RT_distance: The pseudo-RT spacing between adjacent spots (default: 1.0)Example:
For a plate with 4 spots per row and RT_distance=1.0:
After correction and sorting, spectra are ordered by actual spotting sequence.
The command line parameters of this tool are:
DeMeanderize -- Orders the spectra of MALDI spotting plates correctly.
Full documentation: http://www.openms.de/doxygen/nightly/html/TOPP_DeMeanderize.html
Version: 3.6.0-pre-nightly-2025-12-13 Dec 13 2025, 02:53:34, Revision: 86457c9
To cite OpenMS:
+ Pfeuffer, J., Bielow, C., Wein, S. et al.. OpenMS 3 enables reproducible analysis of large-scale mass spec
trometry data. Nat Methods (2024). doi:10.1038/s41592-024-02197-7.
Usage:
DeMeanderize <options>
Options (mandatory options marked with '*'):
-in <mzML-file>* Input experiment file, containing the wrongly sorted spectra. (valid formats:
'mzML')
-out <mzML-file>* Output experiment file with correctly sorted spectra. (valid formats: 'mzML')
-num_spots_per_row <integer> Number of spots in one column, until next row is spotted. (default: '48')
(min: '1')
Common TOPP options:
-ini <file> Use the given TOPP INI file
-threads <n> Sets the number of threads allowed to be used by the TOPP tool (default: '1')
-write_ini <file> Writes the default configuration file
--help Shows options
--helphelp Shows all options (including advanced)
INI file documentation of this tool: